Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
There was a time when researchers had direct access to their home hospital's electronic health record (EHR). Data were only available to researchers for hospital(s) that were affiliated with academic institutions that employed them. In many cases, EHR data and their associated databases could only be accessed onsite. Often, data from ancillary services, such as pharmacy and laboratory data, outside of the hospital, were inaccessible. If the needed data were part of a hospital information system module that had not been created yet, it was considered impossible to access and required manual chart review.
Over the years, researchers built systems and modules to enhance the collection of data for patient care, and eventually fuel additional "secondary data" research. Nationally, biomedical informatics visionaries pointed the way and guided us in how to make research dreams into realities. There were many successes, and as many failures to achieve the vision of researchers having untethered access to EHR and other health data. The eventual result of these trials and tribulations is now a new world of health system-wide data warehouses, secure research enclaves, federated research networks, statewide health information exchanges, all-payer claims databases, and national health data repositories. The term “big data” seems diminutive now that we can leverage trillions points of health data for our research across the nation [1].
Observational Health Research has come of age in terms of data and the tools to conduct research. There is still much work to be done in terms of Health Research Data Governance structures, policies, and procedures. As CODIAC for Health grows and evolves, this chapter will be enhanced to serve as your go-to for the "whys," "whats," and "hows" of it all.
Butte A. . TEDxSanFrancisco. 2017.
Visit other chapters in CODIAC for Health using the or menu in the upper left corner.
Below is an example pipeline or process for conducting research with Health Data such as EHR data. The list is by no means exhaustive. However, it is a good place to start. A page could be written on each of the steps in the pipeline - and most likely will be in future releases of CODIAC for Health.
Conduct a literature review.
Explicitly describe the research question.
Form an interdisciplinary team that can guide and perform each step of the study.
Fully specify the research protocol in advance of executing the study.
Apply for IRB approval of the study.
Apply for an Institutional Reliance Agreement, if necessary.
Execute a Data (Transfer and) Use Agreement (DUA, DTUA), as required
Comply with any application and approval procedures set forth by the data provider.
Request access to / Set up computing infrastructure, as necessary.
Assess the suitability (strengths and weaknesses) of the dataset(s) to be used in the study.
Assess the quality of the dataset(s).
Define the study cohort (and matching cases, if applicable).
Create standard code sets for each clinical concept in the cohort definition and every independent and dependent variable.
Compose a computable data request / data extraction specification.
Clean and stage extracted data for analysis; handle missing values according to protocol.
Characterize the study cohort (and matching cases, if applicable).
Adjust for any bias or confounders in the data.
Analyze the data according to protocol.
Produce research products.
Comply with any review procedures required by the data provider.
Publish your work!
Secondary Analysis of Electronic Health Records [Internet]. Cham (CH): Springer; 2016. Available from: doi: 10.1007/978-3-319-43742-2
Ed. Hulin Wu et al. Statistics and machine learning methods for EHR data: from data extraction to data analytics. CRC Press 2021; ISBN 978-0-367-44239-2
O’Neil ST, Beasley W, Loomba J, Patrick S, Wilkins KJ, Crowley KM., Anzalone, AJ (Eds.) (2023). The Researcher’s Guide to N3C: A National Resource for Analyzing Real-World Health Data. DOI:
Blewett LA, Call KT, Turner J, Hest R. Data Resources for Conducting Health Services and Policy Research. Annu Rev Public Health. 2018 Apr 1;39:437-452. doi: 10.1146/annurev-publhealth-040617-013544. Epub 2017 Dec 22. PMID: 29272166; PMCID: .
Sayers EW, Beck J, Bolton EE, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021 Jan 8;49(D1):D10-D17. doi: 10.1093/nar/gkaa892. PMID: 33095870; PMCID: .
Shang N, Weng C, Hripcsak G. A conceptual framework for evaluating data suitability for observational studies. J Am Med Inform Assoc. 2018 Mar 1;25(3):248-258. doi: 10.1093/jamia/ocx095. PMID:
Weber GM, Mandl KD, Kohane IS. Finding the Missing Link for Big Biomedical Data. JAMA. 2014;311(24):2479–2480. doi:10.1001/jama.2014.4228
Athena Concept Browser (various vocabularies with emphasis on OMOP codes)
ICD10Data (ICD-10-CM)*
CDC ICD Tool (ICD-10-CM)*
ICD9Data (ICD-9-CM)*
SNOMED CT Browsers (SNOMED CT)
Procedure Codes
Athena Concept Browser (various vocabularies with emphasis on OMOP codes)
ICD10Data (ICD-10-PCS)*
(HCPCS)*
(CPT)
Laboratory Codes
Athena Concept Browser (various vocabularies with emphasis on OMOP codes)
SearchLOINC (LOINC)
Medication Codes
Athena Concept Browser (various vocabularies with emphasis on OMOP codes)
RxNav (RxNorm)
(NDC)
*On October 1, 2015, there was a switch from ICD-9-CM to ICD-10-CM. If you are requesting or extracting data from or prior to October 1, 2015, you will need to specify both ICD9 and ICD10 codes.
A clinical concept may have multiple associated codes, each referencing a specific sub-category of that concept. Using pre-gathered and categorized code groupings can help ensure that your study team finds all relevant data. Your study team may benefit from using several of the following methods/resources when looking for code groupings.
Review Methods and Appendix sections of research literature for code groupings
Use the UMLS Metathesaurus Browser which containes terms, codes, hierarchies, definitions, and other relationships and attributes from many vocabularies. These include CPT, ICD-10-CM, LOINC, MeSH, RxNorm, and SNOMED CT.
The OHDSI ATLAS demo contains many code lists (concept sets) across all domains (diagnoses, procedures, medications, etc.). However, many of these concept sets have not been approved by anyone. Those that have been reviewed and approved are prefixed with brackets, “[ ]”, however even these are subject to change. All concept sets taken from ATLAS should be reviewed by a subject matter expert before use.
The National COVID Cohort Collaborative (N3C) has recommended additional diagnosis/problem list code groupings. If you do not have access to the N3C Enclave, you may find a sampling of these codes sets here: .
Refer to the Exercises and Case Studies chapter for examples using following Code Maps and Indexes:
Diagnosis/problem: , ,
Medication: ,
Procedure: ,
"Clinicians and data scientists must apply the same level of academic rigor when analyzing research from clinical databases as they do with more traditional methods of clinical research." [1]
Conducting research with data from the Electronic Health Record (EHR) requires a structured process and a team science approach. The process should include protocols and standards for requesting or extracting the data, assessing the quality of the data, cleaning, standardizing, and analyzing the data, and maintaining security to ensure the confidentiality of the data. A multi-disciplinary team capable of overseeing and performing each specialized step of the process is essential.
-> Research questions, cohorts, and methodologies must be clearly defined using standard clinical definitions and terminologies. Attention must be paid to the timing of exposure and outcome events.
-> Understand the limitations of EHR data. Data may be incomplete, and the quality of the data can vary across sources. Use robust data management strategies to ensure "clean" data and properly handle missing values.
-> Observational studies are susceptible to confounding due to non-random assignment of treatments or exposures. Adjust for confounding factors and be aware of bias that may be inherent in the data.
-> Practice Open Science! Ensure transparency in the study design, data processing, and analysis to promote reproducibility. For NIH-funded studies, comply with the .
-> Practice Team Science! Collaborate with clinicians, biomedical informaticians, biostatisticians, and data scientists to ensure the study is both methodologically sound and clinically meaningful.
"There may come a time when data can be aggregated automatically from multiple EHR environments to answer a particular question without relying on a human to understand the particular idiosyncrasies of each institution’s data and EHR system. Until that day, effective EHR data set analysis requires collaboration with clinicians and scientists who have knowledge of the diseases being studied and the practices of their particular health care systems; informaticians with experience in the underlying structures of biomedical record repositories at their own institutions and the characteristics of their data; data harmonization experts to help with data transformation, standardization, integration, and computability; statisticians and epidemiologists well versed in the limitations and opportunities of EHR data sets and related sources of potential bias; machine learning experts; and at least one expert in regulatory and ethical standards." []
Lokhandwala S, Rush B. Objectives of the Secondary Analysis of Electronic Health Record Data. 2016 Sep 10. In: Secondary Analysis of Electronic Health Records [Internet]. Cham (CH): Springer; 2016. Chapter 1. Available from: / doi: 10.1007/978-3-319-43742-2_1
Kohane IS, Aronow BJ, Avillach P, Beaulieu-Jones BK, Bellazzi R, Bradford RL, Brat GA, Cannataro M, Cimino JJ, García-Barrio N, Gehlenborg N, Ghassemi M, Gutiérrez-Sacristán A, Hanauer DA, Holmes JH, Hong C, Klann JG, Loh NHW, Luo Y, Mandl KD, Daniar M, Moore JH, Murphy SN, Neuraz A, Ngiam KY, Omenn GS, Palmer N, Patel LP, Pedrera-Jiménez M, Sliz P, South AM, Tan ALM, Taylor DM, Taylor BW, Torti C, Vallejos AK, Wagholikar KB; Consortium For Clinical Characterization Of COVID-19 By EHR (4CE); Weber GM, Cai T. What Every Reader Should Know About Studies Using Electronic Health Record Data but May Be Afraid to Ask. J Med Internet Res. 2021 Mar 2;23(3):e22219. doi: 10.2196/22219. PMID: ; PMCID: PMC7927948.
Secondary Analysis of Electronic Health Records [Internet]. Cham (CH): Springer; 2016. Available from: / doi: 10.1007/978-3-319-43742-2
Agniel D, Kohane IS, Weber GM. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ. 2018 Apr 30;361:k1479. doi: 10.1136/bmj.k1479. Erratum in: BMJ. 2018 Oct 18;363:k4416. doi: 10.1136/bmj.k4416. PMID: ; PMCID: PMC5925441.
Callahan A, Shah NH, Chen JH. Research and Reporting Considerations for Observational Studies Using Electronic Health Record Data. Ann Intern Med. 2020 Jun 2;172(11 Suppl):S79-S84. doi: 10.7326/M19-0873. PMID: ; PMCID: PMC7413106.
Shang N, Weng C, Hripcsak G. A conceptual framework for evaluating data suitability for observational studies
The Electronic Health Record (EHR) is a digital version of a patient’s medical history, maintained over time by healthcare providers. It contains detailed information such as demographics, medical history, diagnoses, procedures, medications, and diagnostic test results and images. EHRs can streamline data collection and management, enable high-quality patient care, and facilitate communication between healthcare professionals. The EHR can provide an abundance of real-world data for observational research to study disease progression and treatment effectiveness.
EHRs aggregate data from various sources and systems within a healthcare system and can provide a broad view of patient health.
In extracting data from the EHR, large sample sizes may be obtained at a fraction of the cost of primary data collection.
EHRs can provide a longitudinal record of a patient's health. However, it is important to consider that the entirety of a patient's healthcare interactions may not occur within a single health system.
Variations in EHR systems and standards can hinder data integration across institutions.
EHRs are optimized for collecting and utilizing data in the care of an individual patient. Extracting and staging data for research can be challenging.
As is true of all datasets, incomplete, inconsistent, or inaccurate data can impact the validity of research findings. Research results based on data from a single healthcare system can rarely be directly applied in another setting.
EHRs reflect real-world healthcare settings. As such, the data reflect the clinical practices of the healthcare organization and the population it serves.
Handling sensitive patient information requires strict compliance with privacy, confidentiality, and security policies and procedures.
Secondary Analysis of Electronic Health Records [Internet]. Cham (CH): Springer; 2016. Available from: doi: 10.1007/978-3-319-43742-2.
Ehrenstein V, Kharrazi H, Lehmann H, et al. Obtaining Data From Electronic Health Records. In: Gliklich RE, Leavy MB, Dreyer NA, editors. Tools and Technologies for Registry Interoperability, Registries for Evaluating Patient Outcomes: A User’s Guide, 3rd Edition, Addendum 2 [Internet]. Rockville (MD): Agency for Healthcare Research and Quality (US); 2019 Oct. Available from:
Kohane IS, Aronow BJ, Avillach P, Beaulieu-Jones BK, Bellazzi R, Bradford RL, Brat GA, Cannataro M, Cimino JJ, García-Barrio N, Gehlenborg N, Ghassemi M, Gutiérrez-Sacristán A, Hanauer DA, Holmes JH, Hong C, Klann JG, Loh NHW, Luo Y, Mandl KD, Daniar M, Moore JH, Murphy SN, Neuraz A, Ngiam KY, Omenn GS, Palmer N, Patel LP, Pedrera-Jiménez M, Sliz P, South AM, Tan ALM, Taylor DM, Taylor BW, Torti C, Vallejos AK, Wagholikar KB; Consortium For Clinical Characterization Of COVID-19 By EHR (4CE); Weber GM, Cai T. What Every Reader Should Know About Studies Using Electronic Health Record Data but May Be Afraid to Ask. J Med Internet Res. 2021 Mar 2;23(3):e22219. doi: 10.2196/22219. PMID: 33600347; PMCID: .
The Health Insurance Portability and Accountability Act (HIPAA), Public Law 104-191, enacted on August 21, 1996, protects the privacy of "protected health information" (PHI) [1]. There are 18 elements of PHI as defined by HIPAA:
Names;
All geographical subdivisions smaller than a State, including street address, city, county, precinct, zip code, and their equivalent geocodes, except for the initial three digits of a zip code, if according to the current publicly available data from the Bureau of the Census: (1) The geographic unit formed by combining all zip codes with the same three initial digits contains more than 20,000 people; and (2) The initial three digits of a zip code for all such geographic units containing 20,000 or fewer people is changed to 000;
All elements of dates (except year) for dates directly related to an individual, including birth date, admission date, discharge date, date of death; and all ages over 89 and all elements of dates (including year) indicative of such age, except that such ages and elements may be aggregated into a single category of age 90 or older;
Phone numbers;
Fax numbers;
Electronic mail addresses;
Social Security numbers;
Medical record numbers;
Health plan beneficiary numbers;
Account numbers;
Certificate/license numbers;
Vehicle identifiers and serial numbers, including license plate numbers;
Device identifiers and serial numbers;
Web Universal Resource Locators (URLs);
Internet Protocol (IP) address numbers;
Biometric identifiers, including finger and voice prints;
Full face photographic images and any comparable images; and
Any other unique identifying number, characteristic, or code, except a code to permit re-identification of the de-identified data by the Honest Broker. (Note: this does not include the unique code assigned by an investigator to code the data.)
There are also additional standards and criteria to protect individuals from re-identification. Any code used to replace the identifiers in data sets cannot be derived from any information related to the individual and the master codes, nor can the method to derive the codes be disclosed. For example, a subject’s initials cannot be used to code their data because the initials are derived from their name. Additionally, the researcher must not have actual knowledge that the research subject could be re-identified from the remaining identifiers in the PHI used in the research study. In other words, the information would still be considered identifiable if there was a way to identify the individual even though all of the 18 identifiers were removed.
HIPAA requires that each of the 18 PHI identifiers of the individual or of relatives, employers, or household members of the individual must be removed from medical record information in order for the records to be considered a de-identified “Safe Harbor” dataset.
A dataset can also be de-identified by “expert-determination.” The expert must have professional, academic, or other formal training and experience in using health information de-identification methodologies. The expert may determine that the risk of data re-identification is “very small” when the anticipated recipients use it alone or in combination with other reasonably available information.
To qualify as a Limited Dataset, HIPAA requires that each of the following identifiers of the individual or of relatives, employers, or household members of the individual must be removed from the data.
Names
Postal address information, other than town or city, State, and zip code
Telephone numbers
FAX numbers
U.S. Department of Health and Human Services, Health Information Privacy.[Accessed 2024-11-12];
Institute of Medicine (US) Committee on Health Research and the Privacy of Health Information: The HIPAA Privacy Rule; Nass SJ, Levit LA, Gostin LO, editors. Beyond the HIPAA Privacy Rule: Enhancing Privacy, Improving Health Through Research. Washington (DC): National Academies Press (US); 2009. Available from: / doi: 10.17226/12458
Electronic mail addresses
Social security numbers
Medical record numbers
Health plan beneficiary numbers
Account numbers
Certificate/license numbers
Vehicle identifiers and serial numbers; license plate numbers
Device identifiers and serial numbers
Web Universal Resource Locators (URLs)
Internet Protocol (IP) address numbers
Biometric identifiers
Full face photographic images and any comparable images
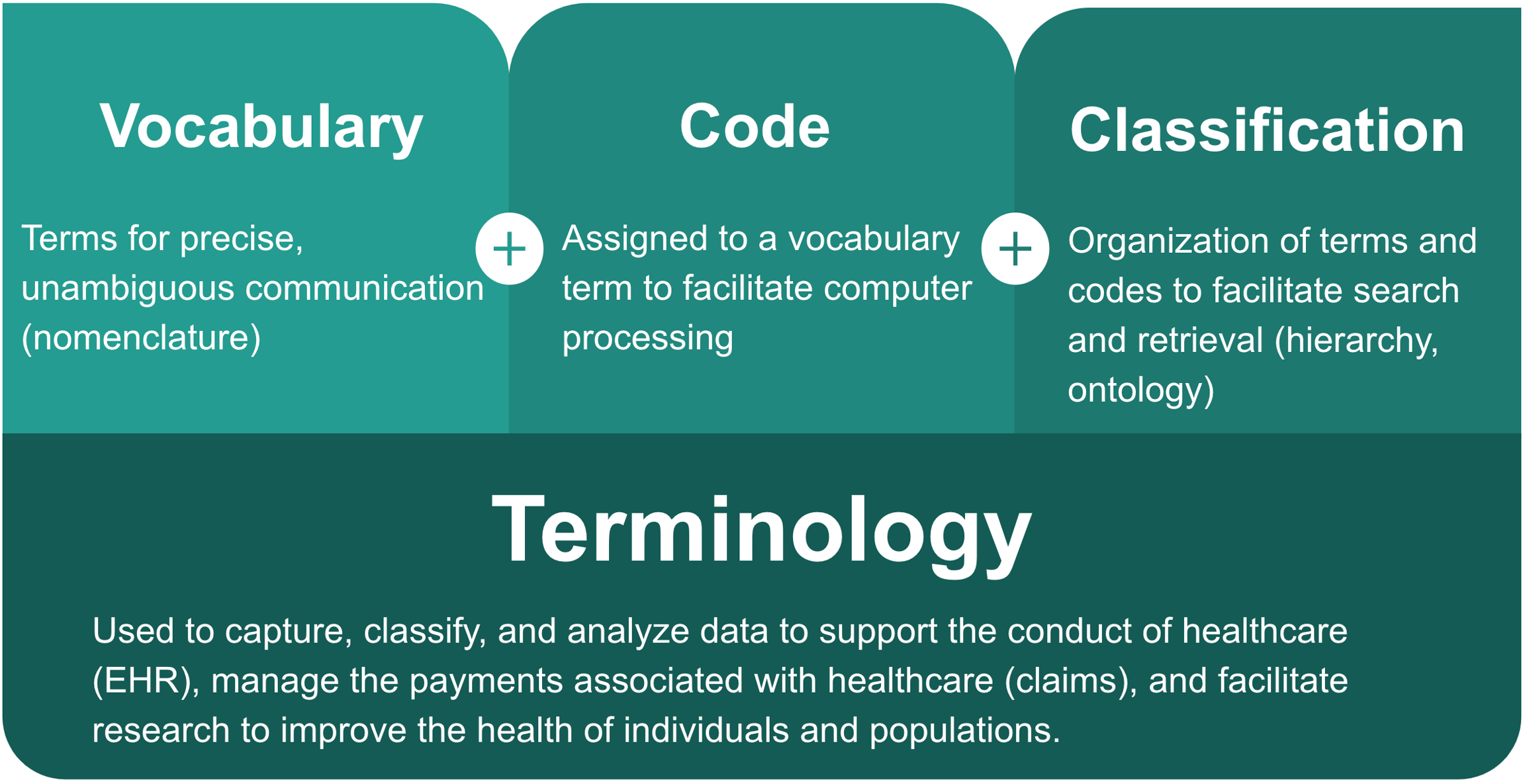
Health terminologies are used to capture, classify, and analyze data to
Support the conduct of health care
Manage the payments associated with healthcare, and
Facilitate research to improve the health of individuals and populations
A health terminology consists of a vocabulary of terms for precise, unambiguous communication, associated codes to facilitate computer processing of those terms, and an organization of those terms and codes to enable search and retrieval.
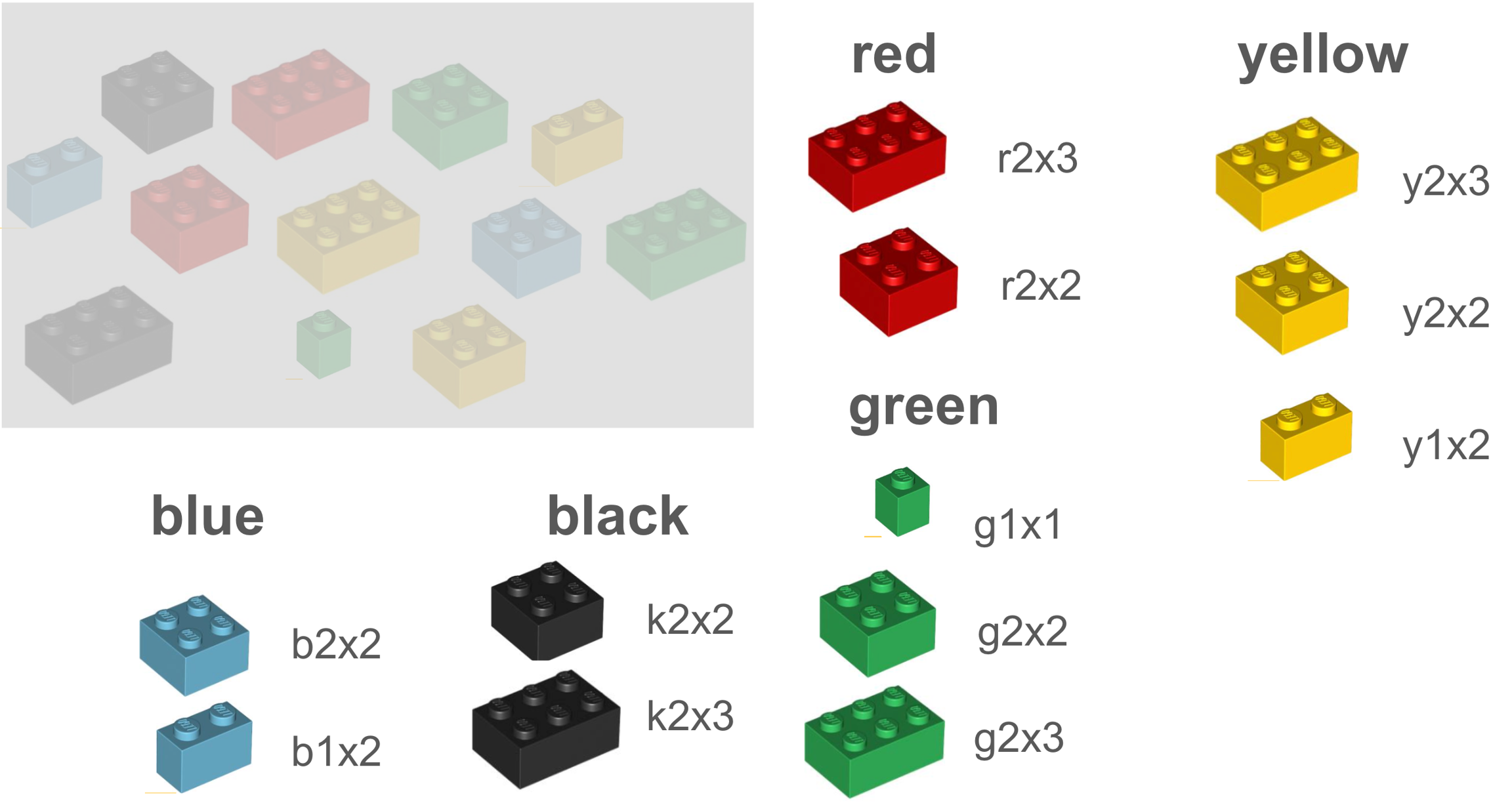
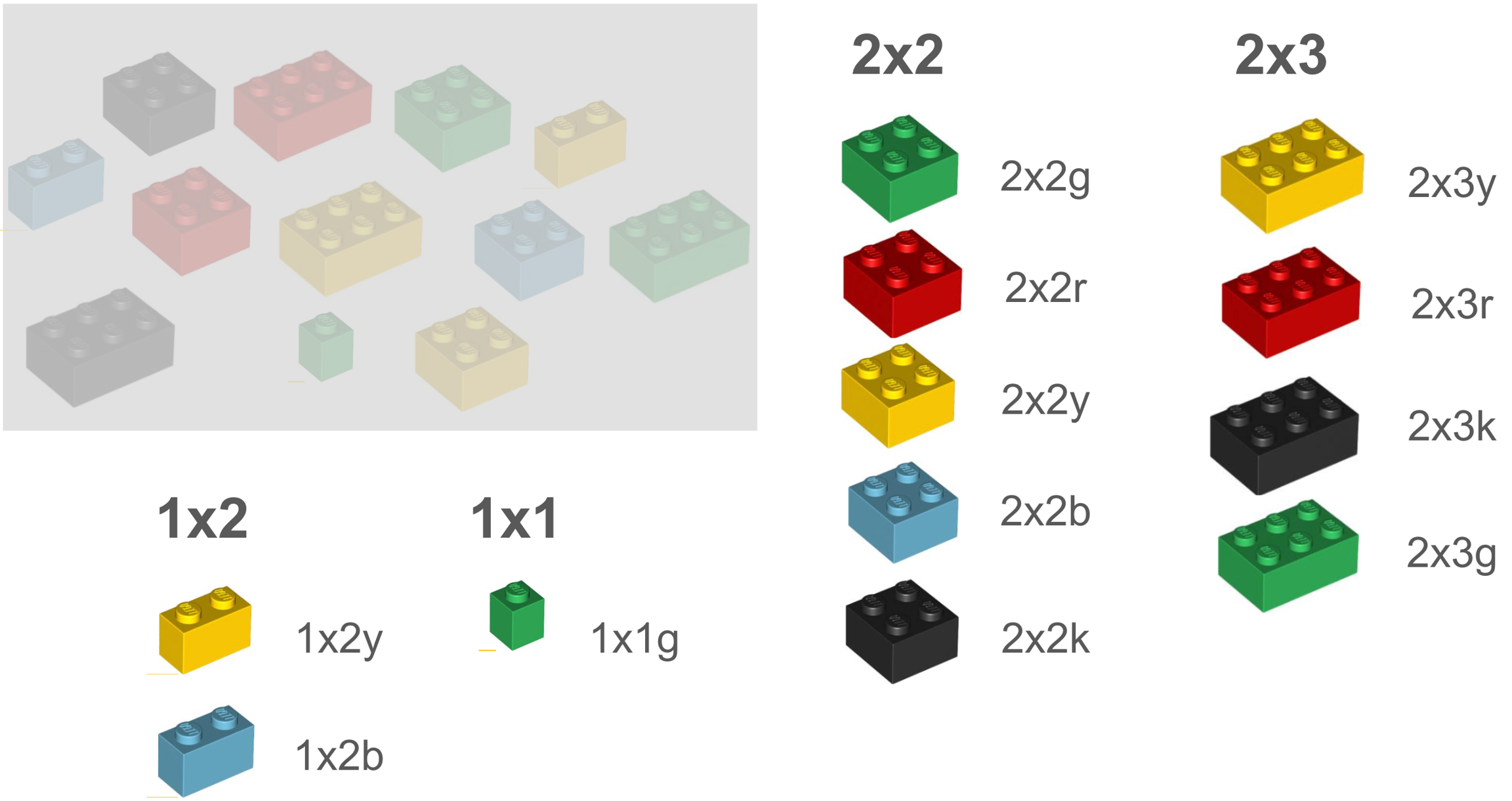
Let’s illustrate a terminology with LEGO® bricks. Each brick represents a vocabulary term. In the below example terminology, the terms are classified or organized by shape or size. And each term is assigned a code.
In another terminology, we may have some of the same or similar terms, but they are classified differently, in this case, by color, leading to the use of different codes.
The following coding systems are the vocabulary standards specified for data elements in the United States Core Data for Interoperability (USCDI), a standardized set of health data classes and constituent data elements for nationwide, interoperable health information exchange. Learn more about USCDI .
"[The International Classification of Diseases Clinical Modification (ICD-CM)] is the official system of assigning codes to diagnoses and procedures associated with hospital utilization in the United States." For more information about ICD-CM, visit .
"The Systematized Medical Nomenclature for Medicine - Clinical Terminology (SNOMED CT) is a clinical terminology sustem that provides a standardized and scientiically validated way of representing clinical inormation captured by clinicians." For more information about SNOMED CT, visit .
"[The Healthcare Common Procedure Coding System (HCPCS)] is a collection of standardized codes that represent medical procedures, supplies, products and services. The codes are used to facilitate the processing of health insurance claims by Medicare and other insurers." For more information about HCPCS, visit .
"The Current Procedural Terminology (CPT) codes offer [clinicians] a uniform language for coding medical services and procedures..." For more information about CPT, visit .
"[Logical Observation Identifiers Names and Codes (LOINC)] is a code system [] for clinical and laboratory observations, health care screening/survey instruments, and document type identifiers." For more information about LOINC, visit .
"RxNorm provides normalized names for clinical drugs and links its names to many of the drug vocabularies commonly used in pharmacy management and drug interaction software..." . For more information about RxNorm, visit .
The FDA requires all clinical drugs to be identified using a National Drug Code (NDC), "which is a universal product identifier for human drugs." For more information about NDC, visit .
The following groupers gather and categorize health terminology codes into searchable groupings. Researchers can efficiently gather health terminology codes through the use of these groupers, utilizing pre-existing phenotypes rather than creating their own.
"Phecodes are widely used and easily adapted phenotypes based on [ICD] codes. The current version of phecodes (v1.2) was designed primarily to study common/complex diseases diagnosed in adults. [PhecodeX] is an expanded version of phecodes, [creating a] more robust representation of the medical phenome for global use in discovery research."
Researchers can use to find pre-gathered and categorized groups of ICD-9-CM and ICD-10-CM codes.
For step-by-step PhecodeX instructions, refer to .
The Clinical Classifications Software (CCS) for ICD-9-CM (CCSR for ICD-10-CM) is a diagnosis and procedure categorization scheme that collapses ICD codes into a smaller number of clinically meaningful categories that are sometimes more useful for presenting descriptive statistics than individual ICD codes.
For more information about CCS and CCSR, refer to the following resources:
For step-by-step CSS/CCSR instructions, refer to .
The Anatomical Therapeutic Chemical (ATC) classification system divides medications into different groups according to the organ or system on which they act and their therapeutic, pharmacological and chemical properties.
For more information about ATC, visit .
For step-by-step ATC instructions, refer to .
Centers for Disease Control and Prevention. (n.d.). International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM). Retrieved May 6, 2024, from
Vuokko, R. (2021). The Systematized Medical Nomenclature for Clinical Care (SNOMED CT): An Overview of a Clinical Terminology for Electronic Health Records. Healthcare Informatics Research, 27(1), 1–6.
National Library of Medicine. (n.d.). HCPCS. U.S. National Library of Medicine. Retrieved May 6, 2024, from
Bodenreider O, Cornet R, Vreeman DJ. Recent Developments in Clinical Terminologies - SNOMED CT, LOINC, and RxNorm. Yearb Med Inform. 2018 Aug;27(1):129-139. doi: 10.1055/s-0038-1667077. Epub 2018 Aug 29. PMID: 30157516; PMCID: .
Wei WQ, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, Cox NJ, Roden DM, Denny JC. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017 Jul 7;12(7):e0175508. doi: 10.1371/journal.pone.0175508. PMID: 28686612; PMCID: .
Megan M Shuey, William W Stead, Ida Aka, April L Barnado, Julie A Bastarache, Elly Brokamp, Meredith Campbell, Robert J Carroll, Jeffrey A Goldstein, Adam Lewis, Beth A Malow, Jonathan D Mosley, Travis Osterman, Dolly A Padovani-Claudio, Andrea Ramirez, Dan M Roden, Bryce A Schuler, Edward Siew, Jennifer Sucre, Isaac Thomsen, Rory J Tinker, Sara Van Driest, Colin Walsh, Jeremy L Warner, Quinn S Wells, Lee Wheless, Lisa Bastarache,
American Medical Association. (n.d.). CPT overview and code approval. AMA Practice Management. Retrieved May 6, 2024, from https://www.ama-assn.org/practice-management/cpt/cpt-overview-and-code-approval
Centers for Medicare & Medicaid Services. (n.d.). LOINC. CMS Measures Management System. Retrieved May 6, 2024, from http://mmshub.cms.gov/measure-lifecycle/measure-specification/specify-code/LOINC
National Library of Medicine. (n.d.). RxNorm. U.S. National Library of Medicine. Retrieved May 6, 2024, from https://www.nlm.nih.gov/research/umls/rxnorm/index.html
U.S. Food and Drug Administration. (n.d.). National Drug Code Database: Background Information. FDA - U.S. Food and Drug Administration. Retrieved May 6, 2024, from https://www.fda.gov/drugs/development-approval-process-drugs/national-drug-code-database-background-information
Megan M Shuey, William W Stead, Ida Aka, April L Barnado, Julie A Bastarache, Elly Brokamp, Meredith Campbell, Robert J Carroll, Jeffrey A Goldstein, Adam Lewis, Beth A Malow, Jonathan D Mosley, Travis Osterman, Dolly A Padovani-Claudio, Andrea Ramirez, Dan M Roden, Bryce A Schuler, Edward Siew, Jennifer Sucre, Isaac Thomsen, Rory J Tinker, Sara Van Driest, Colin Walsh, Jeremy L Warner, Quinn S Wells, Lee Wheless, Lisa Bastarache, Next-generation phenotyping: introducing phecodeX for enhanced discovery research in medical phenomics, Bioinformatics, Volume 39, Issue 11, November 2023, btad655, https://doi.org/10.1093/bioinformatics/btad655
Agency for Healthcare Research and Quality. (n.d.). Clinical Classifications Software (CCS) - Healthcare Cost and Utilization Project (HCUP). Retrieved May 6, 2024, from https://hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp
World Health Organization. (n.d.). ATC Classification. Retrieved May 6, 2024, from https://www.who.int/tools/atc-ddd-toolkit/atc-classification
The Observational Health Data Sciences and Informatics (OHDSI) organization is described here. For the most up-to-date information, please visit ohdsi.org
"The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) is an open community data standard, designed to standardize the structure and content of observational data and to enable efficient analyses that can produce reliable evidence." [1]
To learn more about the OMOP CDM, view this tutorial.
To explore the latest OMOP CDM (v5.4), view the and .
"A central component of the OMOP CDM is the OHDSI standardized vocabularies. The OHDSI vocabularies allow organization and standardization of medical terms to be used across the various clinical domains of the OMOP common data model and enable standardized analytics that leverage the knowledge base when constructing exposure and outcome phenotypes and other features within characterization, population-level effect estimation, and patient-level prediction studies."
OHDSI analysis tools require the use of concept sets and cohort definitions. Concept sets are groups of OMOP concept IDs selected to define the clinical concepts central to a research question. For more information on concept sets, refer to and this on creating concept sets in ATLAS.
A cohort is set of persons who satisfy one or more cohort inclusion criteria for a duration of time. A cohort definition will include the clinical concepts that define the inclusion criteria as well as the temporal logic for cohort entry. For more information on cohorts, refer to and this on ATLAS cohort design.
OHDSI's is a tool to search and and manually map standardized healthcare vocabularies to standard OMOP concept IDs.
ATLAS is a web-based tool developed by the OHDSI community that facilitates the design and execution of analyses on standardized, patient-level, observational data in the OMOP CDM format. As ATLAS is used for analysis of person-level data, an organization will typically install it in a secure environment. OHDSI hosts an instance with synthetic data that can be used for exploration of the tool and training.
HADES is a collection of open-source R packages developed by the OHDSI community that offer functions which can be used together to perform a complete observational study utilizing standardized, patient-level, observational data in the OMOP CDM format.
To learn more about use of OHDSI standards and tools in Rhode Island, please reach out to [email protected]
OHDSI Standardized Data: The OMOP Common Data Model. [Accessed 2025-01-22];
Reich C, Ostropolets A, Ryan P, Rijnbeek P, Schuemie M, Davydov A, Dymshyts D, Hripcsak G. OHDSI Standardized Vocabularies-a large-scale centralized reference ontology for international data harmonization. J Am Med Inform Assoc. 2024 Feb 16;31(3):583-590. doi: 10.1093/jamia/ocad247. PMID: 38175665; PMCID: .
OHDSI Software Tools. [Accessed 2025-01-22];
Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012 Jan-Feb;19(1):54-60. doi: 10.1136/amiajnl-2011-000376. Epub 2011 Oct 28. PMID: 22037893; PMCID: .
Schuemie M, Reps J, Black A, Defalco F, Evans L, Fridgeirsson E, Gilbert JP, Knoll C, Lavallee M, Rao GA, Rijnbeek P, Sadowski K, Sena A, Swerdel J, Williams RD, Suchard M. Health-Analytics Data to Evidence Suite (HADES): Open-Source Software for Observational Research. Stud Health Technol Inform. 2024 Jan 25;310:966-970. doi: 10.3233/SHTI231108. PMID: 38269952; PMCID: .
Schuemie MJ, Ryan PB, Pratt N, Chen R, You SC, Krumholz HM, Madigan D, Hripcsak G, Suchard MA. Principles of Large-scale Evidence Generation and Evaluation across a Network of Databases (LEGEND). J Am Med Inform Assoc. 2020 Aug 1;27(8):1331-1337. doi: 10.1093/jamia/ocaa103. PMID: 32909033; PMCID:

The All of Us Research Program seeks to accelerate health research and medical breakthroughs by collecting and analyzing data from one million or more individuals living in the United States. The program was designed to engage historically underrepresented groups in biomedical research, with an emphasis on community outreach and transparency in research.
Research participants self-selectively enroll and share their data with the program. Data may include electronic health records (EHR), data from wearable devices, physical measurements, surveys, and whole genome sequencing and genotyping arrays. The data are curated and made available within a secure data repository. [1]
Anyone may visit the to learn about the data and explore aggregated participant data with the . Researchers may for access to the Researcher Workbench and conduct research within the secure environment.
Brown University researchers interested in accessing the the All of Us Researcher Workbench should review the for more information.
The CHoRUS project is a data generation project with the goal of creating an ethically-sourced, AI-ready dataset to support future discoveries in clinical care. The project will collect 100,000 patients worth of data centrally from 14 contributor institutions in the United States and combing those data in a cloud repository.
Learn more about CHoRUS on the .
Global.health is an open access global repository and visualization platform for real-time, anonymized, epidemiological line list data for infectious diseases and emerging outbreaks.
Global.health datasets can be viewed within the online or downloaded as a collection of .csv files, categorized by country. Researchers can also view displaying cases across the world.
Learn more about Global.health on the .
Established in 2004, Informatics for Integrating Biology and the Bedside () is an NIH-funded National Center for Biomedical Computing (NCBC) based in Boston, MA. i2b2 seeks to enable clinical researchers to accelerate the translation of genomic and clinical findings into novel diagnostics, prognostics, and therapeutics through the creation of software and a methodological framework.
The i2b2 tranSMART Foundation is "an Open-Source Community Enabling collaboration for precision medicine, through sharing, integration, standardization, and analysis of heterogeneous data from healthcare and research."
Led by the , the Consortium for Clinical Characterization of COVID-19 by EHR () is an international consortium for electronic health record (EHR) data-driven studies of the COVID-19 pandemic. 4CE seeks to inform doctors, epidemiologists, and the public about COVID-19 treatments and patient outcomes.
The National Clinical Cohort Collaborative (N3C) has four enclaves (as of 10/18/2024): N3C COVID, N3C Education, N3C Cancer, and N3C Renal. The Education Enclave provides simulated datasets for researchers to develop and practice the skills needed to analyze real-world data. The Cancer and Renal Enclaves are part of a broader feasibility testing initiative being done to refine the overall governance, data linkage, and institutional partnership components of N3C. These domain-specific enclaves are governed by their data contributors.
"The National COVID Cohort Collaborative (N3C) is a collaboration among the -supported hubs, distributed clinical data networks (, , , ), and other partner organizations, with overall stewardship by NIH’s . The N3C aims to improve the efficiency and accessibility of analyses with COVID-19 clinical data, expand our ability to analyze and understand COVID, and demonstrate a novel approach for collaborative data sharing."
Founded in 2014, the Observational Health Data Sciences and Informatics (OHDSI, pronounced “Odyssey”) is a multi-stakeholder, interdisciplinary, open-science collaborative. OHDSI's mission is "To improve health by empowering a community to collaboratively generate the evidence that promotes better health decisions and better care." OHDSI produces large-scale, real-world analytics through an international network of researchers and observational health databases and a central coordinating center at Columbia University.
For more information about OHDSI, refer to and the .
The Patient-Centered Outcomes Research Institute (PCORI®) is an independent, non-profit research funding organization that funds comparative clinical effectiveness research (CER).
For more information about PCORI, please visit their .
The National Patient-Centered Clinical research Network, PCORnet®, is a distributed network of organizations and standardized health data for patient-centered health research, particularly CER. PCORnet® was developed with funding from the Patient-Centered Outcomes Research Institute® (PCORI®).
For more information, please see their .
The Science Collaborative for Health disparities and Artificial intelligence bias Reduction (ScHARe) is a cloud-based platform for population science including social determinants of health (SDOH), and data sets designed to accelerate research in health disparities, health and healthcare delivery outcomes, and artificial intelligence (AI) bias mitigation strategies.
ScHARe offers a wealth of datasets in the following three categories:
Google-hosted Public Datasets: publicly accessible, federated, de-identified datasets hosted by Google through the Google Cloud Public Dataset Program
ScHARe-hosted Public Datasets: publicly accessible, de-identified datasets hosted by ScHARe
ScHARe-hosted Project Datasets: publicly accessible and controlled-access, funded program/project datasets shared by NIH grantees and intramural investigators to comply with the NIH Data Sharing Policy
Learn more about ScHARe and ScHARe datasets on the .
TriNetX is a global health research network which enables researchers to perform large-scale observational studies with real-world data. Data from electronic health records (EHRs) are aggregated, anonymized, and made available through a secure analytics platform. Through TriNetX, researchers can assess study feasibility and identify eligible patient cohorts. TriNetX is used by pharmaceutical companies and academic and clinical research institutions to perform cohort analyses, conduct comparative effectiveness and epidemiological studies. []
For more information, visit their .
All of Us. [Accessed 2024-11-11];
Chorus for AI. (n.d.). Home. Retrieved May 1, 2024, from
Global.health. (n.d.). About Global.health. Retrieved May 1, 2024, from
O’Neil ST, Beasley W, Loomba J, Patrick S, Wilkins KJ, Crowley KM., Anzalone, AJ (Eds.) (2023). The Researcher’s Guide to N3C: A National Resource for Analyzing Real-World Health Data. DOI:
All of Us Research Program Genomics Investigators. Genomic data in the All of Us Research Program. Nature. 2024 Mar;627(8003):340-346. doi: 10.1038/s41586-023-06957-x. Epub 2024 Feb 19. PMID: ; PMCID: PMC10937371.
Cronin RM, Jerome RN, Mapes B, Andrade R, Johnston R, Ayala J, Schlundt D, Bonnet K, Kripalani S, Goggins K, Wallston KA, Couper MP, Elliott MR, Harris P, Begale M, Munoz F, Lopez-Class M, Cella D, Condon D, AuYoung M, Mazor KM, Mikita S, Manganiello M, Borselli N, Fowler S, Rutter JL, Denny JC, Karlson EW, Ahmedani BK, O'Donnell CJ; Vanderbilt University Medical Center Pilot Team, and the Participant Provided Information Committee. Development of the Initial Surveys for the All of Us Research Program. Epidemiology. 2019 Jul;30(4):597-608. doi: 10.1097/EDE.0000000000001028. PMID: ; PMCID: PMC6548672.
i2b2 tranSMART Foundation. [Accessed 2024-11-11];
4CE. [Accessed 2024-11-11];
N3C. (n.d.). Covid About. [Accessed 2024-10-07];
OHDSI Who We Are. [Accessed 2024-09-30];
OHDSI Mission, Vision & Values. [Accessed 2024-09-30];
Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong IC, Rijnbeek PR, van der Lei J, Pratt N, Norén GN, Li YC, Stang PE, Madigan D, Ryan PB. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574-8. PMID: ; PMCID: PMC4815923.
PCORI. [Accessed 2024-11-11];
PCORnet. [Accessed 2024-11-11];
National Institute on Minority Health and Health Disparities. (n.d.). ScHARe: Science of Health and Racial Equity Program. Retrieved May 1, 2024, from
TriNetX. [Accessed 2024-11-11];
Doerr M, Grayson S, Moore S, Suver C, Wilbanks J, Wagner J. Implementing a universal informed consent process for the All of Us Research Program. Pac Symp Biocomput. 2019;24:427-438. PMID: ; PMCID: PMC6417826.
Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, Payne PRO, Pfaff ER, Robinson PN, Saltz JH, Spratt H, Suver C, Wilbanks J, Wilcox AB, Williams AE, Wu C, Blacketer C, Bradford RL, Cimino JJ, Clark M, Colmenares EW, Francis PA, Gabriel D, Graves A, Hemadri R, Hong SS, Hripscak G, Jiao D, Klann JG, Kostka K, Lee AM, Lehmann HP, Lingrey L, Miller RT, Morris M, Murphy SN, Natarajan K, Palchuk MB, Sheikh U, Solbrig H, Visweswaran S, Walden A, Walters KM, Weber GM, Zhang XT, Zhu RL, Amor B, Girvin AT, Manna A, Qureshi N, Kurilla MG, Michael SG, Portilla LM, Rutter JL, Austin CP, Gersing KR; N3C Consortium. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021 Mar 1;28(3):427-443. doi: 10.1093/jamia/ocaa196. PMID: ; PMCID: PMC7454687.
Suver C, Harper J, Loomba J, Saltz M, Solway J, Anzalone AJ, Walters K, Pfaff E, Walden A, McMurry J, Chute CG, Haendel M. The N3C governance ecosystem: A model socio-technical partnership for the future of collaborative analytics at scale. J Clin Transl Sci. 2023 Nov 14;7(1):e252. doi: 10.1017/cts.2023.681. PMID: ; PMCID: PMC10789985.
Hripcsak G, Schuemie MJ, Madigan D, Ryan PB, Suchard MA. Drawing Reproducible Conclusions from Observational Clinical Data with OHDSI. Yearb Med Inform. 2021 Aug;30(1):283-289. doi: 10.1055/s-0041-1726481. Epub 2021 Apr 21. PMID: ; PMCID: PMC8416226.
Reich C, Ostropolets A, Ryan P, Rijnbeek P, Schuemie M, Davydov A, Dymshyts D, Hripcsak G. OHDSI Standardized Vocabularies-a large-scale centralized reference ontology for international data harmonization. J Am Med Inform Assoc. 2024 Feb 16;31(3):583-590. doi: 10.1093/jamia/ocad247. PMID: ; PMCID: PMC10873827.